Lab 1. Data Wrangling
PUBH 6199: Visualizing Data with R, Summer 2025
2025-05-22
Outline for today
- Introduction to GitHub and Git
- Data transformation
- Data tidying
About GitHub
- GitHub is a cloud-based platform for version control and collaboration.
- Storing your code in a “repository” on GitHub allows you to:
- Track changes to your code over time
- Collaborate with others on projects, including your “future self”
- Share your work with the world
- Made possible by the open-source software, Git

About Git
- Git is a version control system that allows you to track changes to files.
- A typical Git-based workflow includes:
- Clone a repository from GitHub to your local machine
- Branch off the main copy of the files that you are working on
- Edit files independently and safely on your own branch
- Let Git keep track of the changes you and others make
- Let Git intelligently merge your changes back into the main copy of the files

How do Git and GitHub work together?
What is a Git repository?
- A collection of files and their history, can be local (on your computer) or remote (on GitHub)
- When you make changes (or commits) to the files, Git keeps track of the changes
Plenty to do in your browser
- Create a Git repository, create branches, upload and edit files
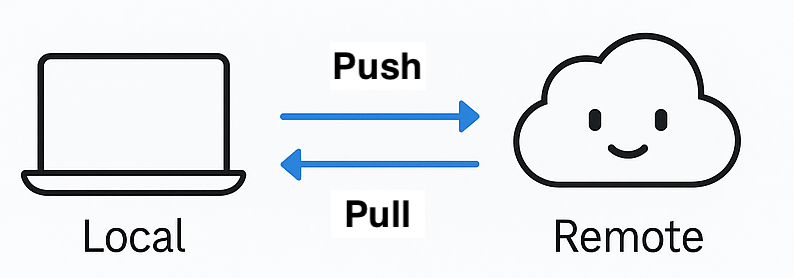
But, most people work locally, then continue to sync local changes with the remote repository on GitHub
- Use Git commands in the terminal or GitHub Desktop
- Pull the latest changes from the remote repository
- Push back your own changes to the same remote repository
![]()
In-Class Activity:
GitHub and RStudio tutorial
Prerequisites
- You have a GitHub account
- You have downloaded and installed Git
- You have downloaded and installed RStudio
5 minutes to catch up on these if you haven’t done so already!
Create the remote repository on GitHub
- Accept the invitation to join the GitHub Classroom, check your email
- Accept the assignment titled lab 1, check your email or use this link https://classroom.github.com/a/XXXX
- Navigate to GitHub, under the class organization, you should see a repository named
lab1-<your-github-username>.
Clone the repository with RStudio
- On GitHub, navigate to the Code tab of the repository
- Click the green <> Codebutton
- Clcik the Copy to clipboard button to copy the repository URL
- Open RStudio on your local environment
- Click File, New Project, Version Control, Git
- Paste the URL you copied from GitHub into the Repository URL field and enter TAB to move to the Project directory name field
- Click Create Project
Edit the lab notebook in RStudio
- In RStudio, click Files, Open File, and select 1-lab1.qmd
- Update the header - put your name in the
authorargument and put today’s date in thedateargument. - Save the file, and click Render to generate the HTML file.
Commit and push the changes to GitHub
- In RStudio, click the Git tab in the upper right pane
- Click Commit
- In the Commit window, check the box next to the file you want to commit (
1-lab1.qmdand1-lab1.html) - Enter a commit message in the Commit message field (e.g., “Update lab notebook header”)
- Click the Commit button
- Click the Pull button to fetch any remote changes
- Click the Push button to push your changes to GitHub
- Navigate to your GitHub repository in your browser and check that the changes have been pushed successfully
Congratulations!

Introducing GitHub Flow

Image by Yan Min Thwin
Create local branches with Git
Tip
You can do these using the Git GUI in RStudio, I am showing you the command line version so you can learn a different method and choose what you prefer.
- In RStudio click the Terminal tab in the lower left pane, next to the Console tab
Note
If you cannot find the Terminal tab, you can also open a terminal window by clicking on the Tools menu and selecting Terminal > New Terminal. If that doesn’t work, check if your RStudio is out of date. Click Help, About RStudio to check the current version.

Create local branches with Git
- In the terminal, type the following command to create a new branch called
feat/clean-data:
Make local changes with Git
In RStudio, open the 1-lab1.qmd file and make some changes to the text.
For example, you can add a new section called “Data Wrangling” and write a few sentences about what tidy data is about.
You can also add a new code chunk to the file and write some R code to load the tidyverse package and read in a CSV file.
After you are satisfied with your changes, save the file and knit the 1-lab1.qmd file to generate the HTML file.
Commit local changes with Git
- Determine your file’s status.
You should see a message that says “On branch feat/clean-data” and “Changes not staged for commit”.
- Add the changes to the staging area.
- See your file’s current status.
Your files should be listed under Changes to be committed.
- Commit the changes with a message. Replace
with a log message describing the changes.
Open a pull request on GitHub
- Push the changes to the remote repository, replace
with the name of your branch, in this case feat/clean-data
- Navigate to your GitHub repository in your browser
- Click the Compare & pull requests button, if you don’t see it, navigate to the “Pull requests” tab and click the New pull reques button.
- In the “Open a pull request” page, enter a title and description for your pull request. You can add a reviewer, for example your teammate on this pull request.
Merge your pull request on GitHub
Note
Since this is your repository, you probably don’t have anyone to collaborate with (yet). Go ahead and merge your Pull Request now. Later in the semester you may want your teammate to look over your code before they merge.
- On GitHub, navigate to the Pull Request that you just opened.
- Scroll down and click the big green Merge Pull Request button.
- Click Confirm Merge.
- Delete the branch
.
Reference: GitHub and RStudio
Take a Break
~ This is the end of part 1 ~
Outline for today
- Introduction to GitHub and Git
- Data transformation
- Data tidying
“80% of data scientists’ time is spent on data wrangling”
Data wrangling: also known as data cleaning or data preparation, is the process of collecting, cleaning, transforming and organizing data from one “raw” form into another format with the intent of making it more appropriate for analysis.

Source: R for Data Science
Manipulate data in R using dplyr
Commonality:
- The first argument is always a data frame
- The subsequent arguments are the columns of the data frame (without quotes)
- The output is a new data frame . . .
Individuality:
Rows
filter(): filter rowsarrange(): change the order of rowsdistinct(): remove duplicate rows
Columns
select(): select columnsrename(): rename columnsmutate(): add new columns
Groups
group_by(): group rows by one or more columnssummarize(): summarize data by groupsslice_*(): extract specific rowsungroup(): remove grouping
A word on pipe
%>% in {magrittr} or |> in base R
- Pipe is a tool to combine multiple verbs.
- It takes the thing on the left and passes it to the function on the right.
x |> f(y)is equivalent tof(x, y)x |> f(y) |> g(z)is equivalent tog(f(x, y), z).- Pronounces as “then”
- Add pipe to your code using keyboard shortcut Ctrl/Cmd + Shift + M
Lots of verbs to remember!

Refer to this cheat sheet
Practice makes perfect!
~ Head over to lab1 notebook! ~
Outline for today
- Introduction to GitHub and Git
- Data transformation
- Data tidying
Introduction to tidy data
“Happy families are all alike; every unhappy family is unhappy in its own way.”
- Leo Tolstoy, Anna Karenina
“Tidy datasets are all alike, but every messy dataset is messy in its own way.”
- Hadley Wickham, Tidy Data

What is tidy data?

What is an example of untidy data?

Multiple tables, not machine-readable
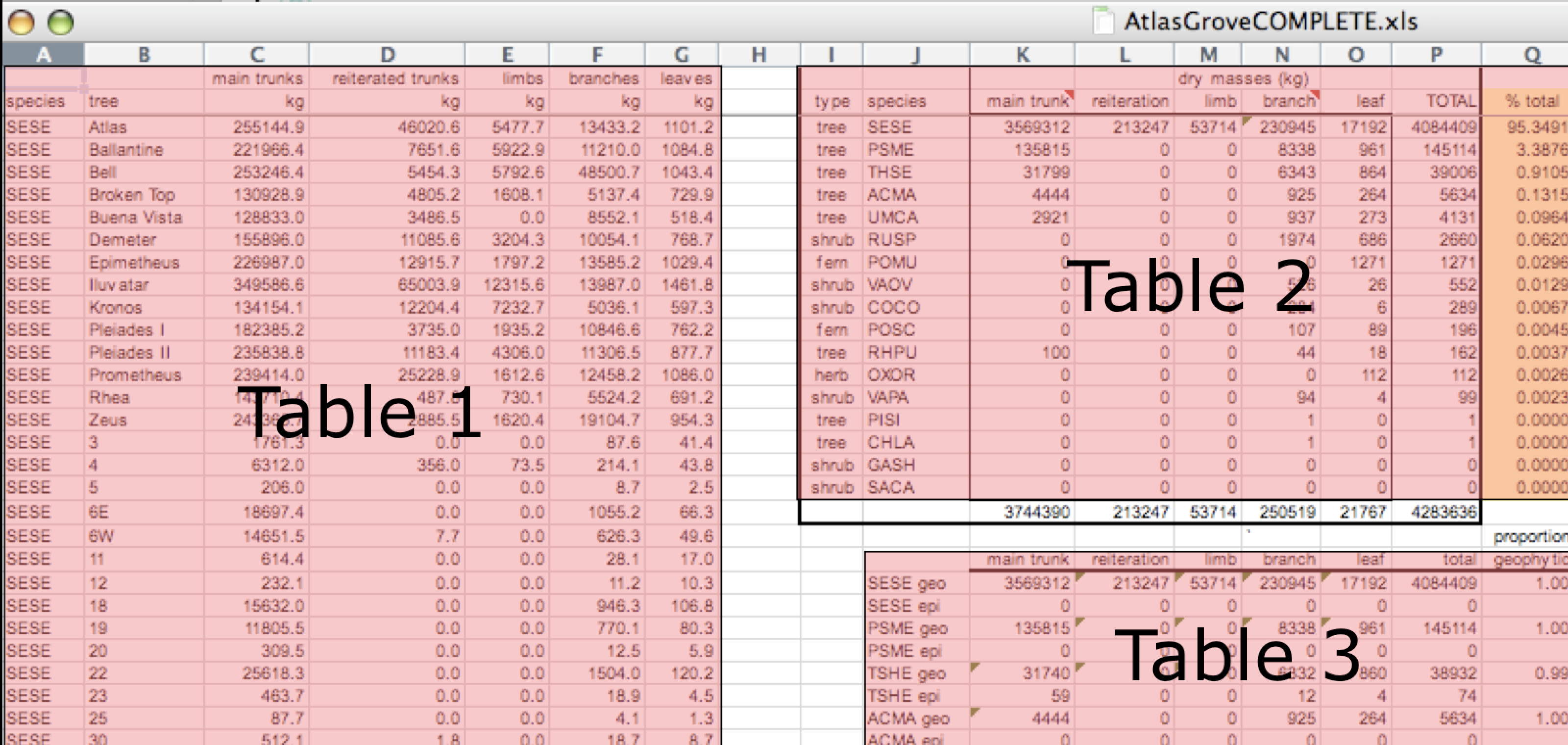
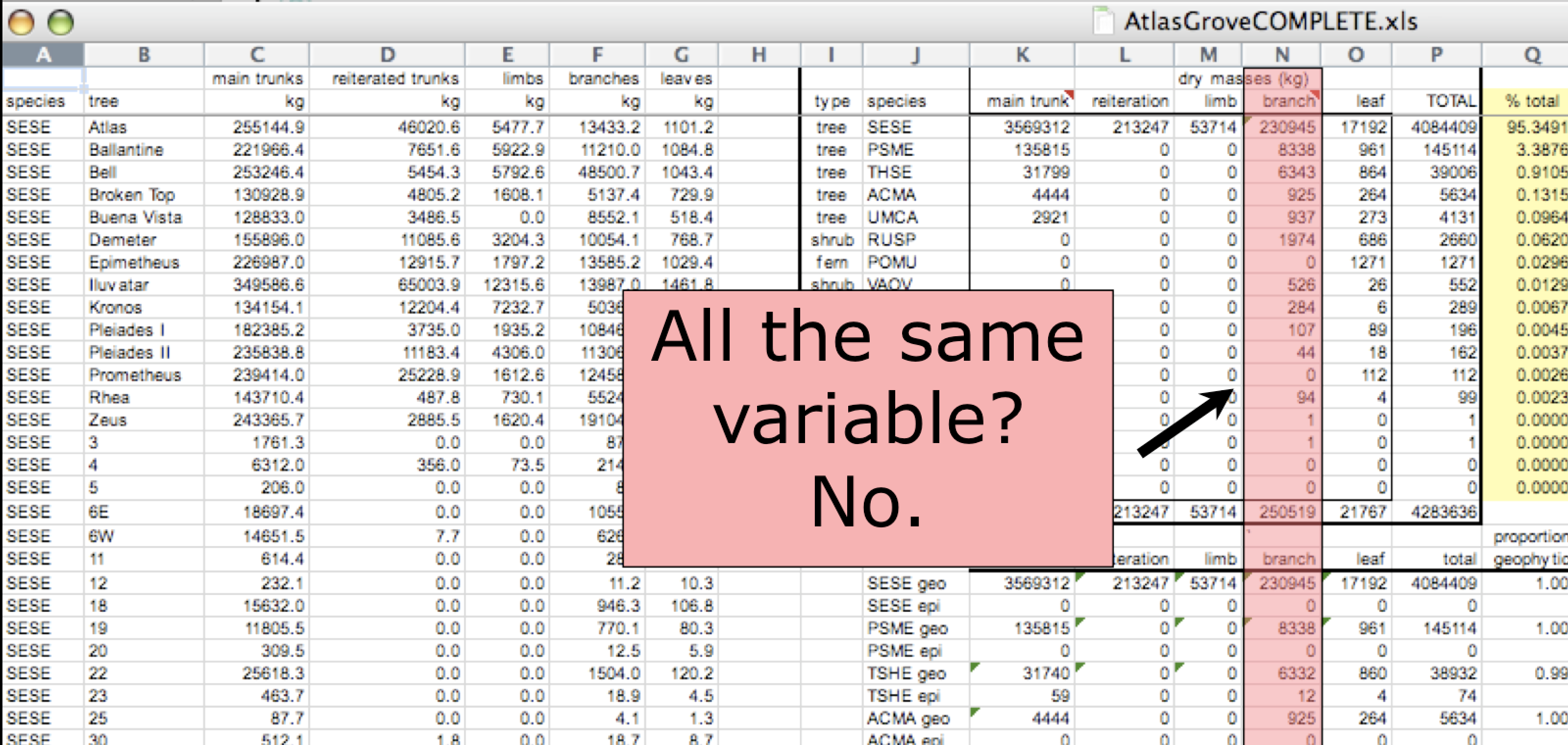
Inconsistent columns
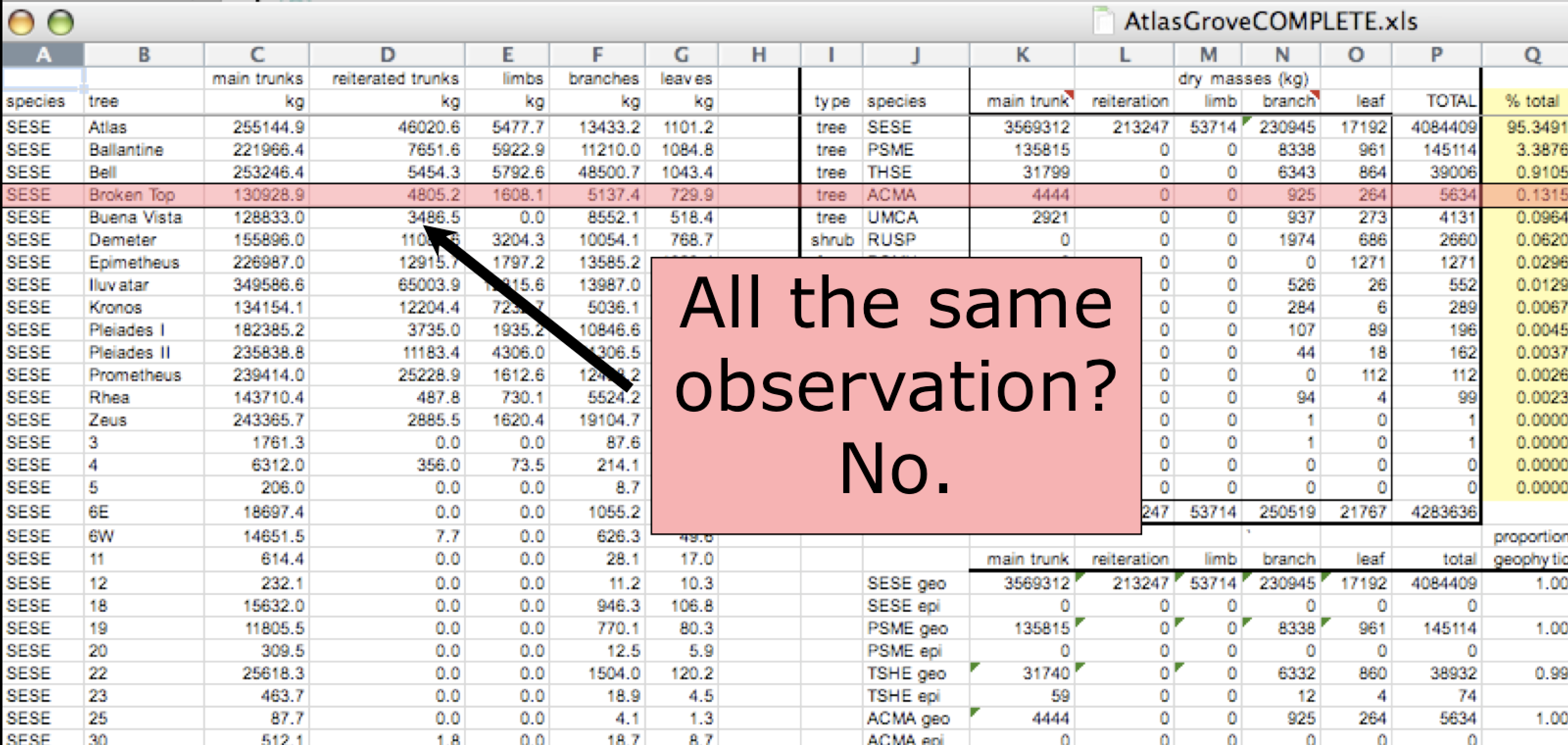
Inconsistent rows
Marginal sums and statistics
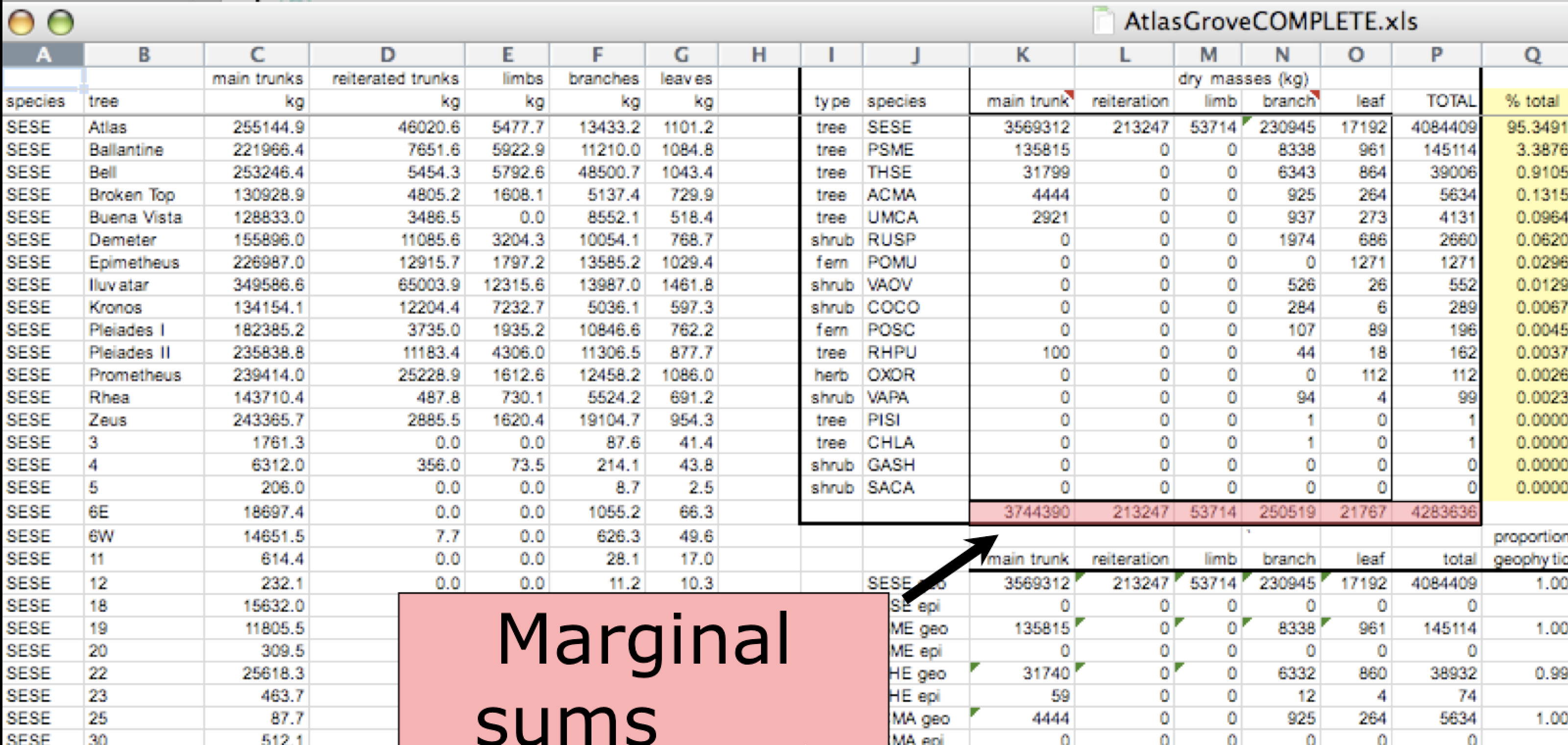
A single untidy table, climate_raw
| date | city | zone | temp_morning | temp_afternoon | humid_morning | humid_afternoon |
|---|---|---|---|---|---|---|
| 2022-07-01 | Phoenix | urban | 79 | 106 | 23 | 39 |
| 2022-07-02 | Phoenix | urban | 85 | 102 | 72 | 71 |
| 2022-07-03 | Phoenix | urban | 77 | 96 | 27 | 57 |
| 2022-07-04 | Phoenix | urban | 83 | 107 | 49 | 56 |
| 2022-07-05 | Phoenix | urban | 78 | 106 | 69 | 41 |
| 2022-07-01 | Miami | coastal | 85 | 110 | 72 | 51 |
| 2022-07-02 | Miami | coastal | 82 | 101 | 54 | 53 |
| 2022-07-03 | Miami | coastal | 81 | 106 | 90 | 63 |
| 2022-07-04 | Miami | coastal | 77 | 100 | 87 | 32 |
| 2022-07-05 | Miami | coastal | 75 | 105 | 73 | 53 |
In-Class Activity:
In pairs, discuss the following:
- What makes
climate_rawuntidy? - Sketch out on paper what a tidy version of
climate_rawwould look like.
Why do untidy data exist and what to do about it?
- Data is collected in a way that is convenient for the collector, not the analyst
- Most people aren’t familiar with the principles of tidy data unless you are a data professional
- To tidy data:
- Begin by figuring out what are the variables and observations
- Talk to the data curator if needed
- pivot your data into a tidy form
pivot_longer()
Suppose we have three patients with ids A, B, and C. Each patient has two blood pressure measurements: bp1 and bp2. The data is in wide format:
We want our new dataset to have three variables: id (already exists), measurement (the column names), and value (the cell values). To achieve this, we pivot df longer:
How does pivot_longer() work?
Repeat id twice
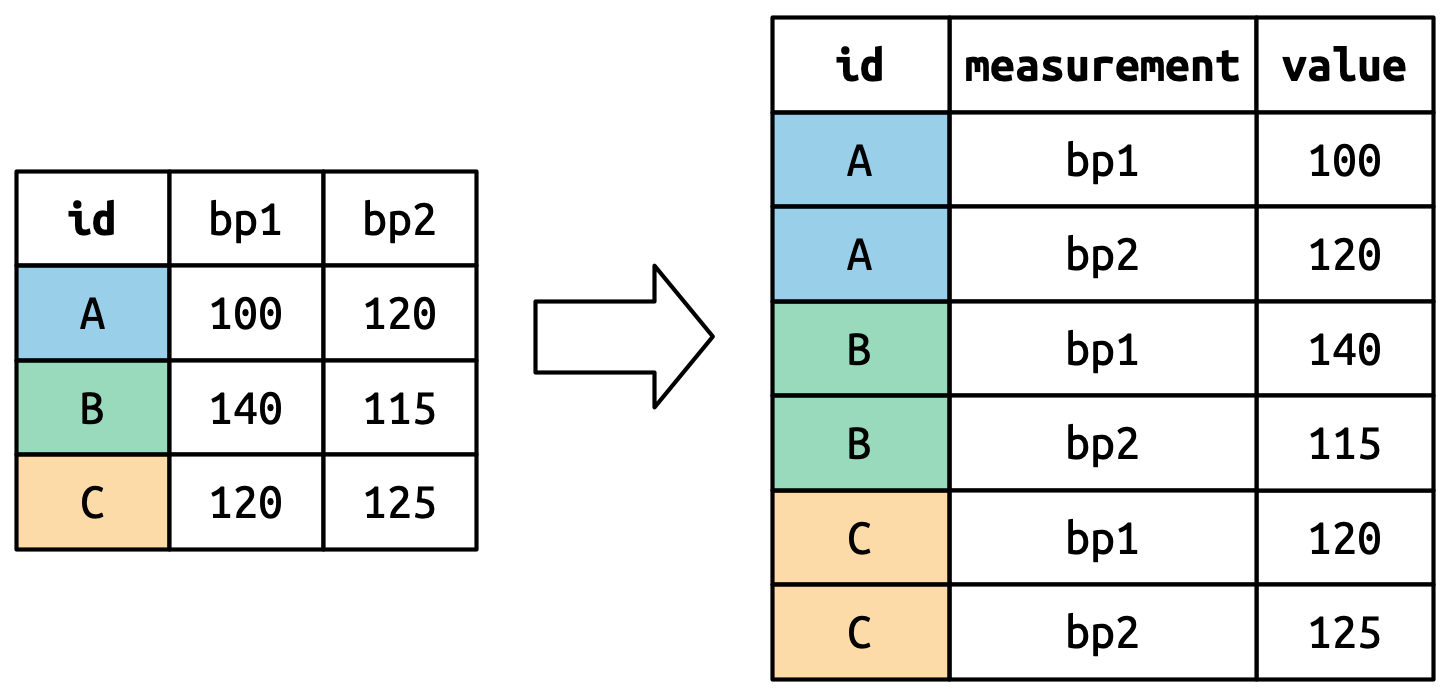
bp1 and bp2 become values in a new column
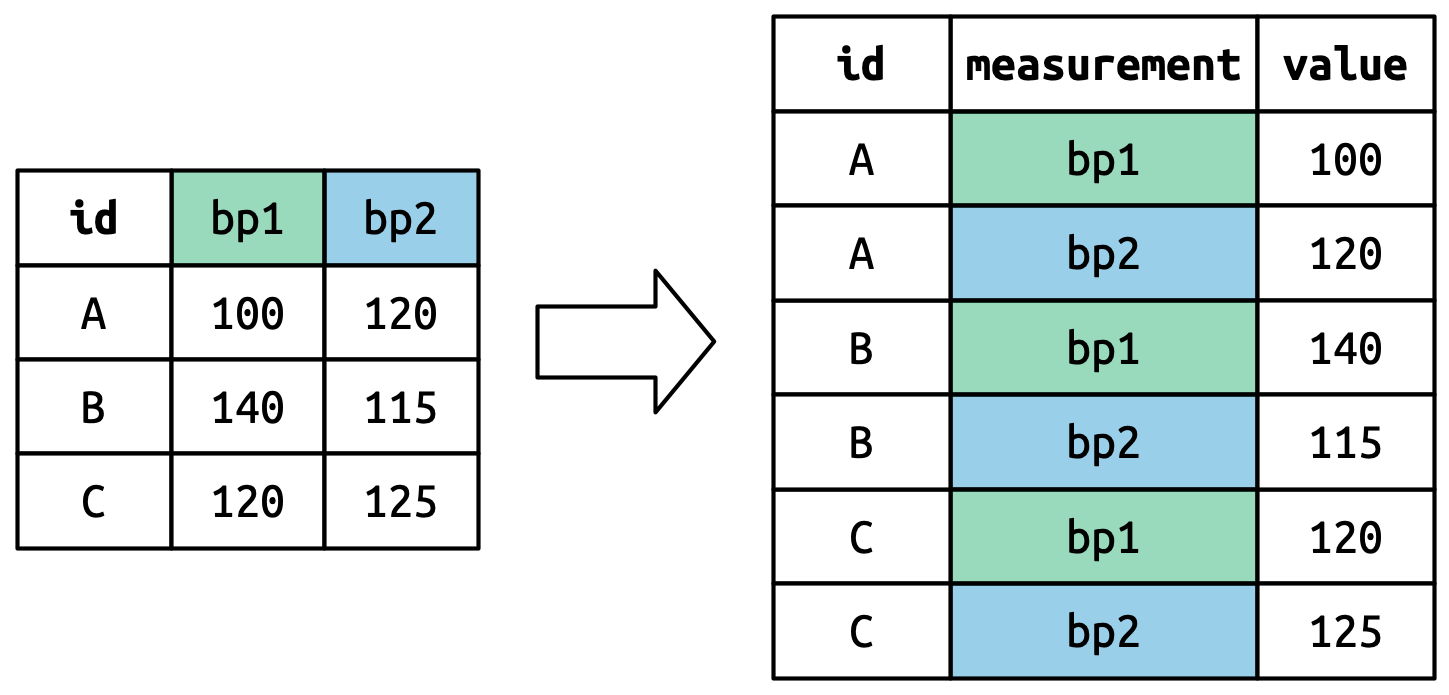
The number of values is preserved and unwound row-by-row.
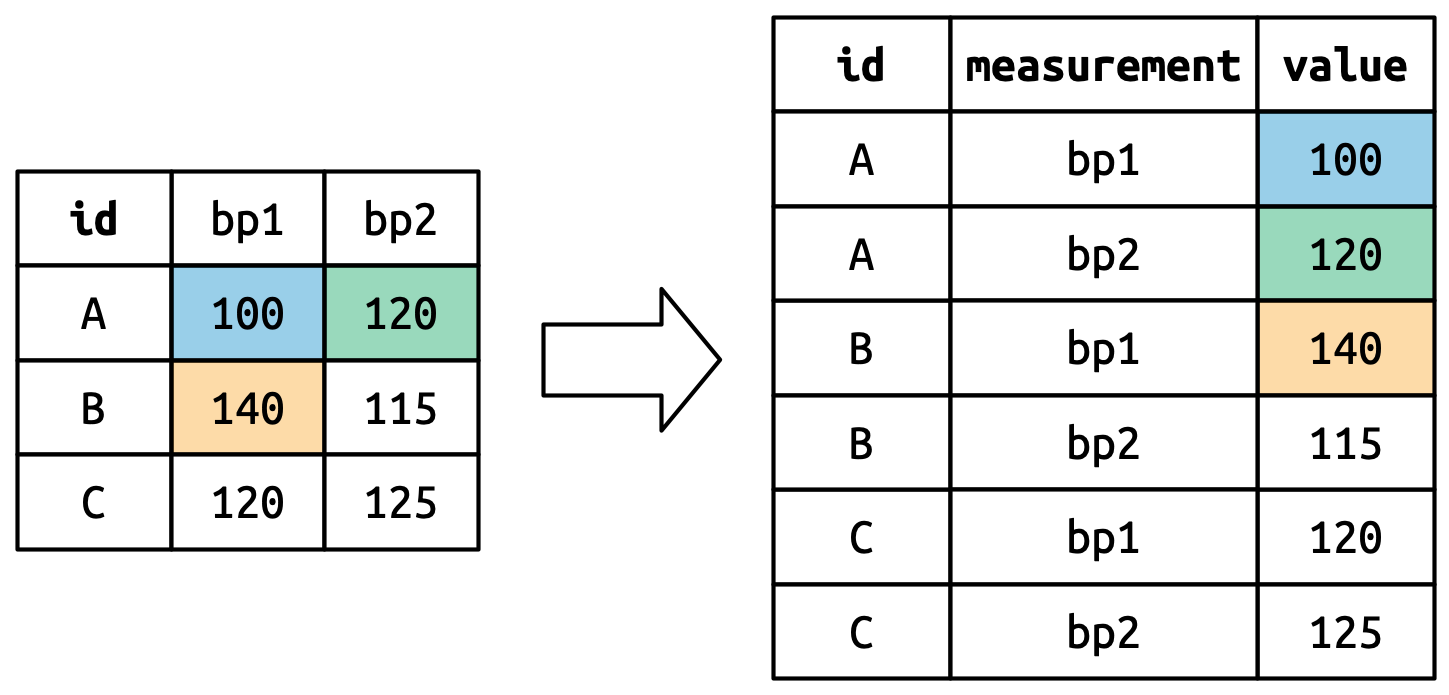
pivot_wider()
Suppose we have two patients with ids A and B. We have three blood measurements on patient A and two on patient B. The data is in long format:
How does pivot_wider() work?
First, figure out what will be the new column names, taken from measurement.
pivot_wider() then combine the columns and rows to generate an empty data frame, then fill it with value in the input.
# A tibble: 2 × 4
id bp1 bp2 bp3
<chr> <lgl> <lgl> <lgl>
1 A NA NA NA
2 B NA NA NA pivot_wider() can make missing values.
Why do we need pivot_wider()?
Isn’t tidy data long?
- Yes — tidy data often means long format, especially for:
- plotting
- filtering
- grouping
- But tidy ≠ always long!
Tidy = Structure
- Each variable in a column, each observation in a row
- Sometimes wide format is tidy — it depends on context.
When do we need pivot_wider()?
- ✅ For modeling:
lm(bp1 ~ bp2)needs one column per variable
- ✅ For presentation:
- Easier to read tables with 1 row per subject
- ✅ For joining:
- Merge with spatial data or metadata
- ✅ To undo a pivot_longer()
Let’s tidy climate_raw
~ Head over to lab1 notebook! ~
End-of-Class Survey
Fill out the end-of-class survey
~ This is the end of Lab 1 ~
PUBH 6199: Visualizing Data with R